About
Molecular Center of Health and Disease Overview
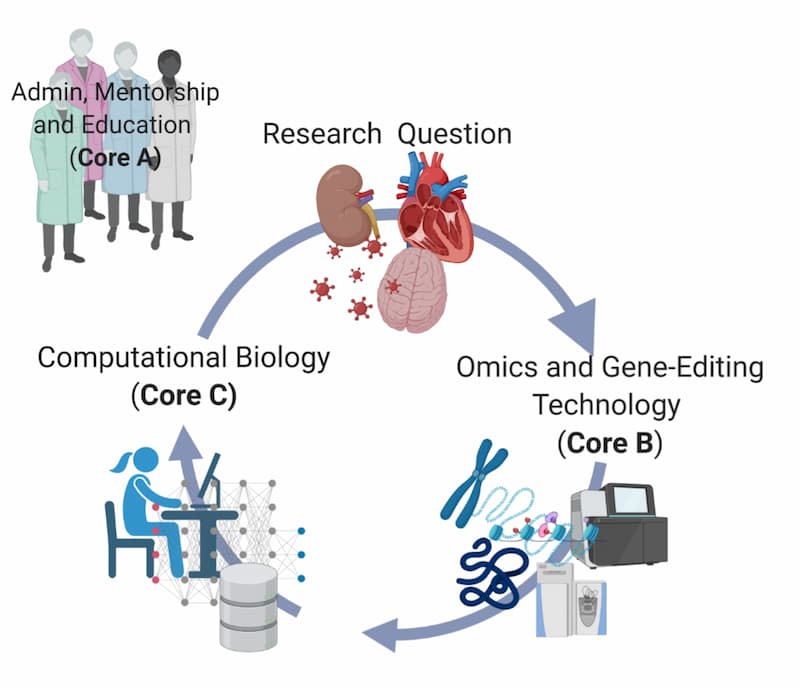
The Molecular Center of Health and Disease (MCHD) is comprised of multiple assorted components, including an administrative unit, mentoring and education program, two research cores, three major research project leaders (RPL) as well as a Pilot Project Program.
The MCHD brings together established and productive basic and clinician researchers from varied backgrounds and expertise to accomplish the Center’s mission. Each Core Director is led by a senior experienced researcher with administrative experience, with other Core Leads having relevant expertise and background to provide RPL and pilot project investigators quality services.
It is the goal of the MCHD to seamlessly integrate each of the below core functions for RPL and pilot project investigators to maximize their full potential to become independent, extramurally funded researchers, that will contribute back to the MCHD.
Core A: Administrative, Mentoring, and Education provides overall leadership for development of infrastructure, education programs, and research core facilities to promote the application of systems biology/omics technology important to human health problems.
Core B: Omics and Gene-Editing provides MCHD investigators access to state-of-the-art omics technologies through several components. It will also serve as a nucleus for collaborative work with other IDeA programs at UMMC as it will offer capabilities that are not currently provided through those Centers
Core C: Research Computing, Bioinformatics, and Biostatistics establishes a high-performance computing resource (both internal and using cloud platforms) for custom analysis pipelines to analyze single omics datasets as well as the integration of multiple omics datasets at UMMC.


